Department of Animal Ecology, Lund University, SE-223 62 Lund, Sweden
Present address:



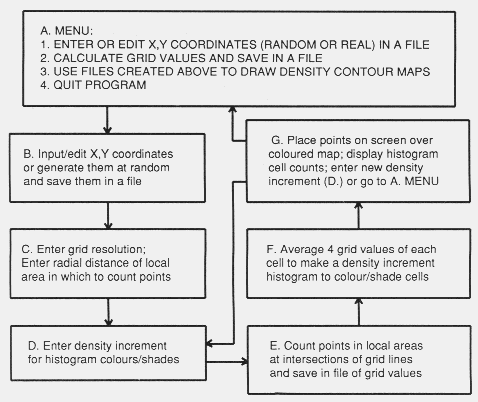
 (1)
(1) (2)
(2) (3)
(3)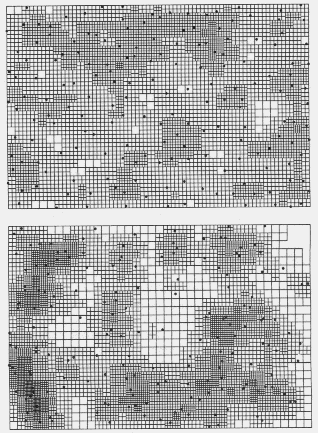
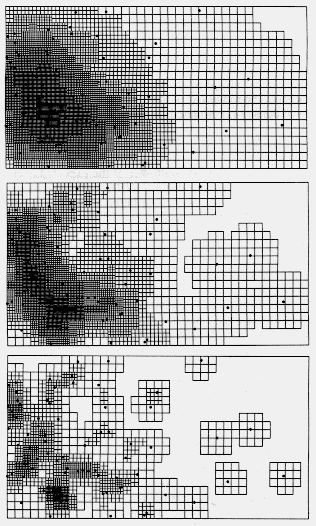
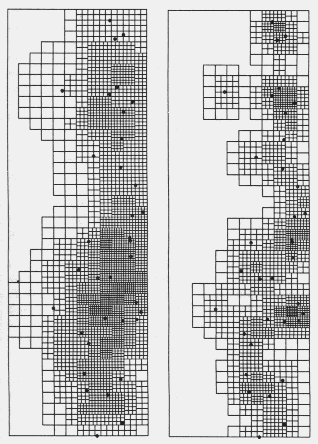
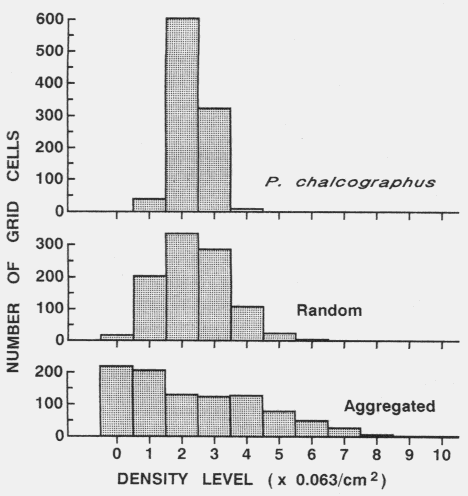
| Table 1. P values (and degrees of freedom) for Chi-square comparisons of grid cell colour frequency histograms from contour maps of Pityogenes chalcographus attacks or random points (data in Fig. 2) with an average histogram from three random data sets. Comparisons were made between histograms from maps constructed at various D (density class increments) and R (expected average nearest neighhour distance) values, while densities and grid parameters were identical. | |||
| P values for Chi-square (df) | |||
|---|---|---|---|
| D = 1 | D = 2 | D = 3 | |
| Random points | |||
| R = 1 | 0.64(3) | 0.54(2) | 0.27(1) |
| R = 2 | 0.80(8) | 0.41(4) | 0.46(3) |
| R = 3 | 0.28(11) | 0.17(6) | 0.08(4) |
| P. chalcographus | |||
| R = 1 | < 0.001(2) | < 0.001(1) | < 0.001(1) |
| R = 2 | < 0.001(4) | < 0.001(2) | < 0.001(1) |
| R = 3 | < 0.001(6) | < 0.001(3) | < 0.001(2) |
|
John A. Byers Department of Animal Ecology, Lund University, SE-223 62 Lund, Sweden Present address: 
|
|---|